Uncertainties
Contents
5. Uncertainties#
Import Python libraries
from __future__ import annotations
import itertools
import json
import logging
import os
import tarfile
from collections import defaultdict
from textwrap import wrap
import jax.numpy as jnp
import matplotlib.pyplot as plt
import numpy as np
import plotly.graph_objects as go
import sympy as sp
import yaml
from ampform.kinematics.phasespace import is_within_phasespace
from ampform.sympy import PoolSum
from IPython.display import Latex, Markdown
from matplotlib import cm
from matplotlib.ticker import FuncFormatter, MultipleLocator
from plotly.subplots import make_subplots
from scipy.interpolate import griddata
from tensorwaves.function.sympy import create_function
from tensorwaves.interface import DataSample, ParametrizedFunction
from tqdm.auto import tqdm
from polarimetry import formulate_polarimetry
from polarimetry.amplitude import AmplitudeModel
from polarimetry.data import (
create_data_transformer,
generate_meshgrid_sample,
generate_phasespace_sample,
)
from polarimetry.function import integrate_intensity, sub_intensity
from polarimetry.io import (
export_polarimetry_field,
mute_jax_warnings,
perform_cached_doit,
perform_cached_lambdify,
)
from polarimetry.lhcb import (
ParameterBootstrap,
load_model_builder,
load_model_parameters,
)
from polarimetry.lhcb.particle import load_particles
from polarimetry.plot import add_watermark, use_mpl_latex_fonts
logging.getLogger("polarimetry.io").setLevel(logging.INFO)
mute_jax_warnings()
FUNCTION_CACHE: dict[sp.Expr, ParametrizedFunction] = {}
UNFOLDED_POOLSUM_CACHE: dict[PoolSum, sp.Expr] = {}
NO_TQDM = "EXECUTE_NB" in os.environ
if NO_TQDM:
logging.getLogger().setLevel(logging.ERROR)
logging.getLogger("polarimetry.io").setLevel(logging.ERROR)
5.1. Model loading#
Formulate models
model_file = "../data/model-definitions.yaml"
particles = load_particles("../data/particle-definitions.yaml")
reference_subsystem = 1
with open(model_file) as f:
model_titles = list(yaml.safe_load(f))
models = {}
for title in tqdm(model_titles, desc="Formulating models", disable=NO_TQDM):
amplitude_builder = load_model_builder(model_file, particles, model_id=title)
model = amplitude_builder.formulate(reference_subsystem)
imported_parameter_values = load_model_parameters(
model_file, model.decay, title, particles
)
model.parameter_defaults.update(imported_parameter_values)
models[title] = model
Unfold symbolic expressions
def unfold_poolsum(expr: PoolSum) -> sp.Expr:
unfolded_expr = UNFOLDED_POOLSUM_CACHE.get(expr)
if unfolded_expr is None:
unfolded_expr = expr.doit()
UNFOLDED_POOLSUM_CACHE[expr] = unfolded_expr
return unfolded_expr
return unfolded_expr
nominal_model_title = "Default amplitude model"
nominal_model = models[nominal_model_title]
unfolded_exprs = {}
for title, model in tqdm(
models.items(), desc="Unfolding expressions", disable=NO_TQDM
):
amplitude_builder = load_model_builder(model_file, particles, model_id=title)
polarimetry_exprs = formulate_polarimetry(amplitude_builder, reference_subsystem)
folded_exprs = {f"alpha_{i}": expr for i, expr in zip("xyz", polarimetry_exprs)}
folded_exprs["intensity"] = model.intensity
unfolded_exprs[title] = {
key: perform_cached_doit(unfold_poolsum(expr).xreplace(model.amplitudes))
for key, expr in tqdm(
folded_exprs.items(), disable=NO_TQDM, leave=False, postfix=title
)
}
Identify unique expressions
expression_hashes = {
title: hash(exprs["intensity"]) for title, exprs in unfolded_exprs.items()
}
table = R"""
$$
\begin{array}{rllr}
&& \textbf{Model description} & n\textbf{ ops.} \\
\hline
"""
for i, (title, expressions) in enumerate(unfolded_exprs.items()):
expr = expressions["intensity"]
h = hash(expr)
for j, v in enumerate(expression_hashes.values()):
if h == v:
break
same_as = ""
if i != j:
same_as = f"= {j}"
ops = sp.count_ops(expr)
title = "".join(Rf"\text{{{t}}}\\ " for t in wrap(title, width=56))
title = Rf"\begin{{array}}{{l}}{title}\end{{array}}"
title = title.replace("^", R"\^{}")
table += Rf" \mathbf{{{i}}} & {same_as} & {title} & {ops:,} \\ \hline" "\n"
table += R"""\end{array}
$$
"""
n_models = len(models)
n_unique_hashes = len(set(expression_hashes.values()))
src = Rf"""
Of the {n_models} models, there are {n_unique_hashes} with a unique expression tree.
"""
if NO_TQDM:
src += "\n:::{dropdown} Show number of mathematical operations per model\n"
src += table
if NO_TQDM:
src += "\n:::"
Markdown(src.strip())
Of the 18 models, there are 9 with a unique expression tree.
Show number of mathematical operations per model
Convert to numerical functions
def cached_lambdify(expr: sp.Expr, model: AmplitudeModel) -> ParametrizedFunction:
func = FUNCTION_CACHE.get(expr)
if func is None:
func = perform_cached_lambdify(
expr,
parameters=model.parameter_defaults,
backend="jax",
)
FUNCTION_CACHE[expr] = func
str_parameters = {str(k): v for k, v in model.parameter_defaults.items()}
func.update_parameters(str_parameters)
return func
jax_functions = {}
original_parameters: dict[str, dict[str, complex | float | int]] = {}
progress_bar = tqdm(desc="Lambdifying to JAX", disable=NO_TQDM, total=len(models))
for title, model in models.items():
progress_bar.set_postfix_str(title)
jax_functions[title] = {
key: cached_lambdify(expr, model)
for key, expr in unfolded_exprs[title].items()
}
original_parameters[title] = dict(jax_functions[title]["intensity"].parameters)
progress_bar.update()
progress_bar.set_postfix_str("")
progress_bar.close()
5.2. Statistical uncertainties#
5.2.1. Parameter bootstrapping#
n_bootstraps = 100
nominal_functions = jax_functions[nominal_model_title]
bootstrap = ParameterBootstrap(model_file, nominal_model.decay, nominal_model_title)
bootstrap_parameters = bootstrap.create_distribution(n_bootstraps, seed=0)
n_events = 100_000
transformer = create_data_transformer(nominal_model)
phsp_sample = generate_phasespace_sample(nominal_model.decay, n_events, seed=0)
phsp_sample = transformer(phsp_sample)
Compute polarimeter field and intensities (statistics & systematics)
resonances = [chain.resonance for chain in nominal_model.decay.chains]
nominal_parameters = dict(original_parameters[nominal_model_title])
stat_grids = defaultdict(list)
stat_decay_rates = defaultdict(list)
for i in tqdm(
range(n_bootstraps),
desc="Computing polarimetry and intensities for parameter combinations",
disable=NO_TQDM,
):
new_parameters = {k: v[i] for k, v in bootstrap_parameters.items()}
for key, func in nominal_functions.items():
func.update_parameters(nominal_parameters)
func.update_parameters(new_parameters)
stat_grids[key].append(func(phsp_sample).real)
I_tot = integrate_intensity(stat_grids["intensity"][-1])
for resonance in resonances:
res_filter = resonance.name.replace("(", R"\(").replace(")", R"\)")
I_sub = sub_intensity(
nominal_functions["intensity"], phsp_sample, [res_filter]
)
stat_decay_rates[resonance.name].append(I_sub / I_tot)
stat_intensities = jnp.array(stat_grids["intensity"])
stat_polarimetry = jnp.array([stat_grids[f"alpha_{i}"] for i in "xyz"])
stat_polarimetry_norm = jnp.sqrt(jnp.sum(stat_polarimetry**2, axis=0))
stat_decay_rates = {k: jnp.array(v) for k, v in stat_decay_rates.items()}
5.2.2. Mean and standard deviations#
assert stat_intensities.shape == (n_bootstraps, n_events)
assert stat_polarimetry.shape == (3, n_bootstraps, n_events)
assert stat_polarimetry_norm.shape == (n_bootstraps, n_events)
n_bootstraps, n_events
(100, 100000)
Compute statistical measures (mean, std, etc.)
stat_alpha_mean = [
jnp.mean(stat_polarimetry_norm, axis=0),
*jnp.mean(stat_polarimetry, axis=1),
]
stat_alpha_std = [
jnp.std(stat_polarimetry_norm, axis=0),
*jnp.std(stat_polarimetry, axis=1),
]
stat_alpha_times_I_mean = [
jnp.mean(stat_polarimetry_norm * stat_intensities, axis=0),
*jnp.mean(stat_polarimetry * stat_intensities, axis=1),
]
stat_alpha_times_I_std = [
jnp.std(stat_polarimetry_norm * stat_intensities, axis=0),
*jnp.std(stat_polarimetry * stat_intensities, axis=1),
]
stat_alpha_times_I_mean = jnp.array(stat_alpha_times_I_mean)
stat_alpha_times_I_std = jnp.array(stat_alpha_times_I_std)
stat_intensity_mean = jnp.mean(stat_intensities, axis=0)
stat_intensity_std = jnp.std(stat_intensities, axis=0)
5.2.3. Distributions#
Define grid for plotting
def interpolate_to_grid(values: np.ndarray, method: str = "linear"):
return griddata(POINTS, values, (X, Y))
resolution = 200
POINTS = np.transpose(
[
phsp_sample["sigma1"],
phsp_sample["sigma2"],
]
)
grid_sample = generate_meshgrid_sample(nominal_model.decay, resolution)
X = np.array(grid_sample["sigma1"])
Y = np.array(grid_sample["sigma2"])
Code for indicating Dalitz region
def create_contour(phsp: DataSample) -> jnp.ndarray:
# See also https://compwa-org.rtfd.io/report/017.html
m0, m1, m2, m3, s1, s2 = sp.symbols("m(:4) sigma(1:3)", nonnegative=True)
filter_expr = is_within_phasespace(
s1,
s2,
nominal_model.parameter_defaults[m0],
nominal_model.parameter_defaults[m1],
nominal_model.parameter_defaults[m2],
nominal_model.parameter_defaults[m3],
outside_value=0,
).doit()
filter_func = create_function(filter_expr, backend="jax")
return filter_func(contour_sample) # noqa: F821
def draw_dalitz_contour(
ax, color: str = "black", width: float = 0.1, **kwargs
) -> None:
ax.contour(
X_CONTOUR,
Y_CONTOUR,
Z_CONTOUR,
colors=color,
linewidths=width,
**kwargs,
)
contour_sample = generate_meshgrid_sample(nominal_model.decay, resolution=500)
X_CONTOUR = np.array(contour_sample["sigma1"])
Y_CONTOUR = np.array(contour_sample["sigma2"])
Z_CONTOUR = create_contour(contour_sample)
del contour_sample, create_contour
Show code cell source
%config InlineBackend.figure_formats = ['png']
plt.rcdefaults()
use_mpl_latex_fonts()
plt.rc("font", size=18)
fig, axes = plt.subplots(
dpi=200,
figsize=(16.7, 8),
gridspec_kw={"width_ratios": [1, 1, 1, 1.18]},
ncols=4,
nrows=2,
sharex=True,
sharey=True,
)
plt.subplots_adjust(hspace=0.02, wspace=0.02)
fig.suptitle(R"Polarimetry field $\vec\alpha$ (statistical \& systematic)")
s1_label = R"$m^2\left(K^-\pi^+\right)$ [GeV$^2$]"
s2_label = R"$m^2\left(pK^-\right)$ [GeV$^2$]"
s3_label = R"$m^2\left(p\pi^+\right)$ [GeV$^2$]"
axes[0, 0].set_ylabel(s2_label)
axes[1, 0].set_ylabel(s2_label)
global_max_std = max(map(jnp.nanmax, stat_alpha_std))
for i in range(4):
if i != 0:
title = Rf"$\alpha_{'xyz'[i-1]}$"
else:
title = R"$\left|\vec\alpha\right|$"
axes[0, i].set_title(title)
draw_dalitz_contour(axes[0, i], zorder=10)
Z = interpolate_to_grid(stat_alpha_mean[i])
mesh = axes[0, i].pcolormesh(X, Y, Z, cmap=cm.RdYlGn_r)
mesh.set_clim(vmin=-1, vmax=+1)
if axes[0, i] is axes[0, -1]:
c_bar = fig.colorbar(mesh, ax=axes[0, i], pad=0.01)
c_bar.ax.set_ylabel(Rf"$\alpha$ averaged with {n_bootstraps} bootstraps")
c_bar.ax.set_yticks([-1, 0, +1])
c_bar.ax.set_yticklabels(["-1", "0", "+1"])
draw_dalitz_contour(axes[1, i], zorder=10)
Z = interpolate_to_grid(stat_alpha_std[i])
mesh = axes[1, i].pcolormesh(X, Y, Z)
mesh.set_clim(vmin=0, vmax=global_max_std)
axes[1, i].set_xlabel(s1_label)
if axes[1, i] is axes[1, -1]:
c_bar = fig.colorbar(mesh, ax=axes[1, i], pad=0.01)
c_bar.ax.set_ylabel("standard deviation")
plt.show()
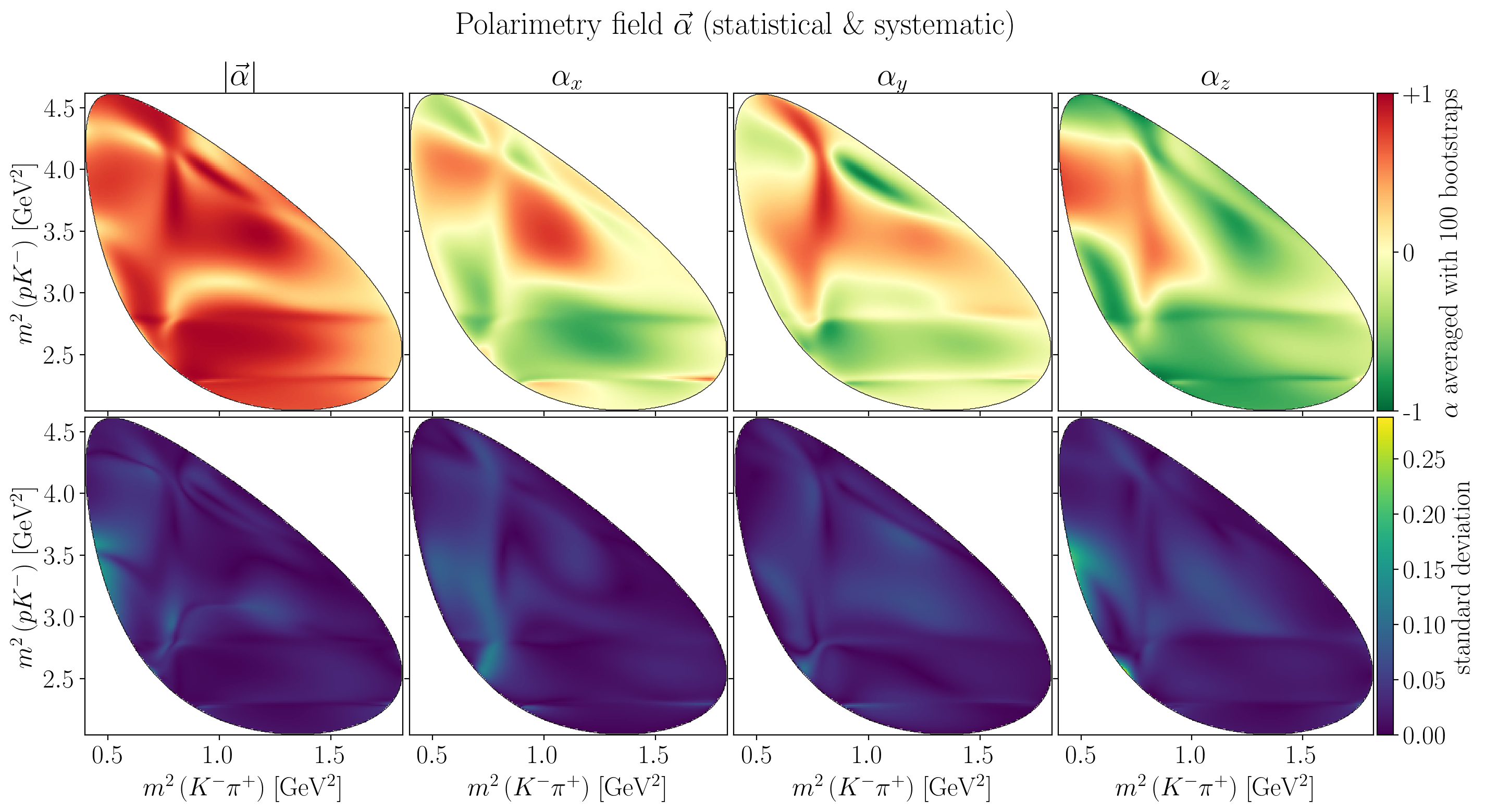
Show code cell source
%config InlineBackend.figure_formats = ['png']
fig, axes = plt.subplots(
dpi=200,
figsize=(16.7, 8),
gridspec_kw={"width_ratios": [1, 1, 1, 1.18]},
ncols=4,
nrows=2,
sharex=True,
sharey=True,
)
plt.subplots_adjust(hspace=0.02, wspace=0.02)
fig.suptitle(R"$\vec\alpha \cdot I$ distributions (statistical \& systematic)")
axes[0, 0].set_ylabel(s2_label)
axes[1, 0].set_ylabel(s2_label)
global_max_mean = jnp.nanmax(jnp.abs(stat_alpha_times_I_mean))
global_max_std = jnp.nanmax(stat_alpha_times_I_std / stat_intensity_mean)
for i in range(4):
if i != 0:
title = Rf"$\alpha_{'xyz'[i-1]}$"
else:
title = R"$\left|\vec\alpha\right|$"
axes[0, i].set_title(title)
axes[1, i].set_xlabel(s1_label)
draw_dalitz_contour(axes[0, i], zorder=10)
Z = interpolate_to_grid(stat_alpha_times_I_mean[i])
mesh = axes[0, i].pcolormesh(X, Y, Z, cmap=cm.RdYlGn_r)
mesh.set_clim(vmin=-global_max_mean, vmax=+global_max_mean)
if axes[0, i] is axes[0, -1]:
c_bar = fig.colorbar(mesh, ax=axes[0, i], pad=0.01)
c_bar.ax.set_ylabel(
Rf"$\alpha \cdot I$ averaged with {n_bootstraps} bootstraps"
)
draw_dalitz_contour(axes[1, i], zorder=10)
Z = interpolate_to_grid(stat_alpha_times_I_std[i] / stat_intensity_mean)
mesh = axes[1, i].pcolormesh(X, Y, Z)
mesh.set_clim(vmin=0, vmax=global_max_std)
if axes[1, i] is axes[1, -1]:
c_bar = fig.colorbar(mesh, ax=axes[1, i], pad=0.01)
c_bar.ax.set_ylabel("standard deviation / intensity")
plt.show()
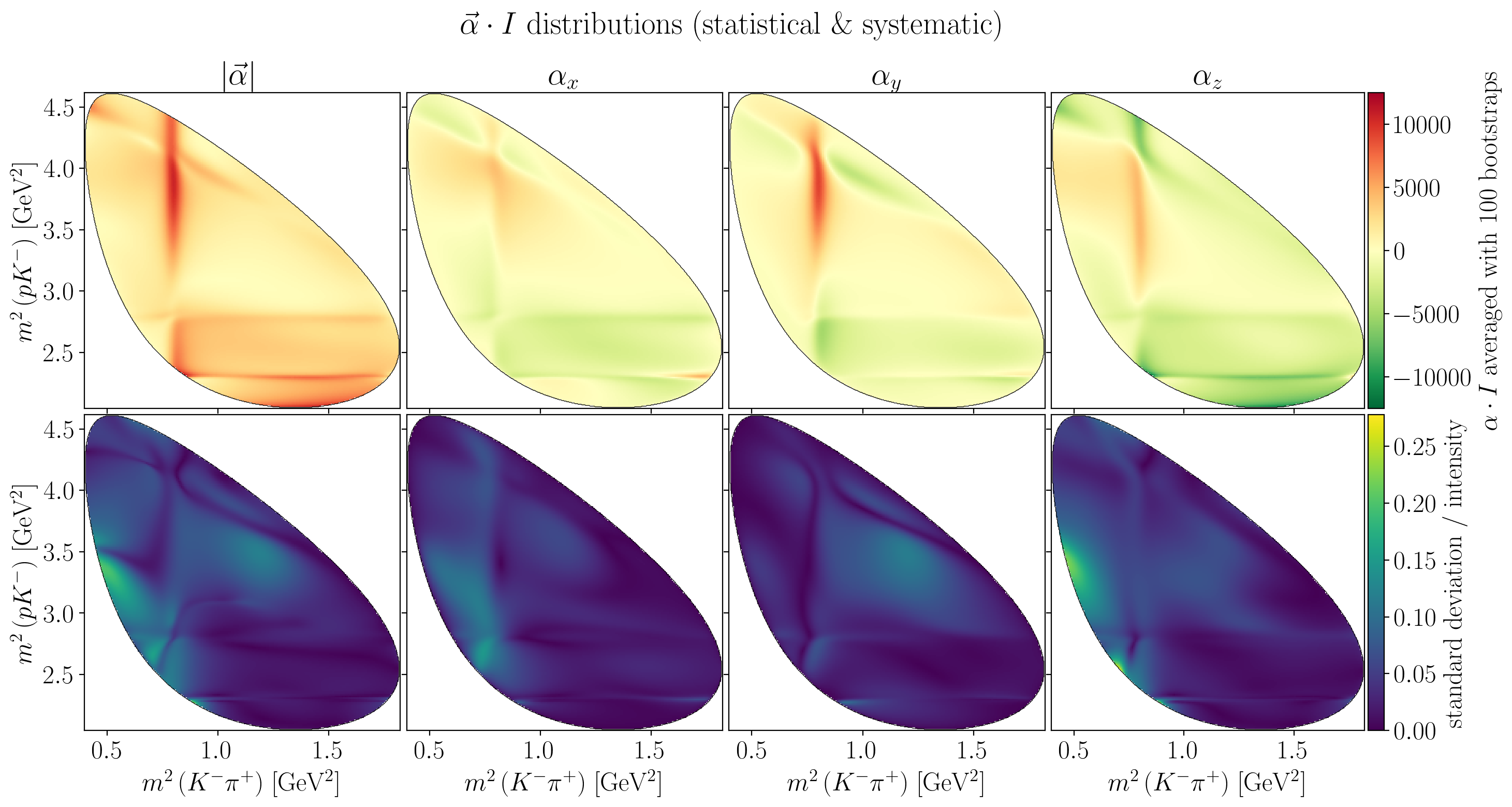
Show code cell source
%config InlineBackend.figure_formats = ['png']
fig, (ax1, ax2) = plt.subplots(
dpi=200,
figsize=(12, 6.2),
ncols=2,
sharey=True,
)
fig.suptitle(R"Intensity distribution (statistical \& systematics)", y=0.95)
ax1.set_xlabel(s1_label)
ax2.set_xlabel(s1_label)
ax1.set_ylabel(s2_label)
Z = interpolate_to_grid(stat_intensity_mean)
mesh = ax1.pcolormesh(X, Y, Z, cmap=cm.Reds)
fig.colorbar(mesh, ax=ax1, pad=0.01)
draw_dalitz_contour(ax1, width=0.2)
ax1.set_title(f"average of {n_bootstraps} bootstraps")
Z = interpolate_to_grid(stat_intensity_std / stat_intensity_mean)
mesh = ax2.pcolormesh(X, Y, Z)
fig.colorbar(mesh, ax=ax2, pad=0.01)
draw_dalitz_contour(ax2, width=0.2)
ax2.set_title("standard deviation / intensity")
fig.tight_layout()
plt.show()
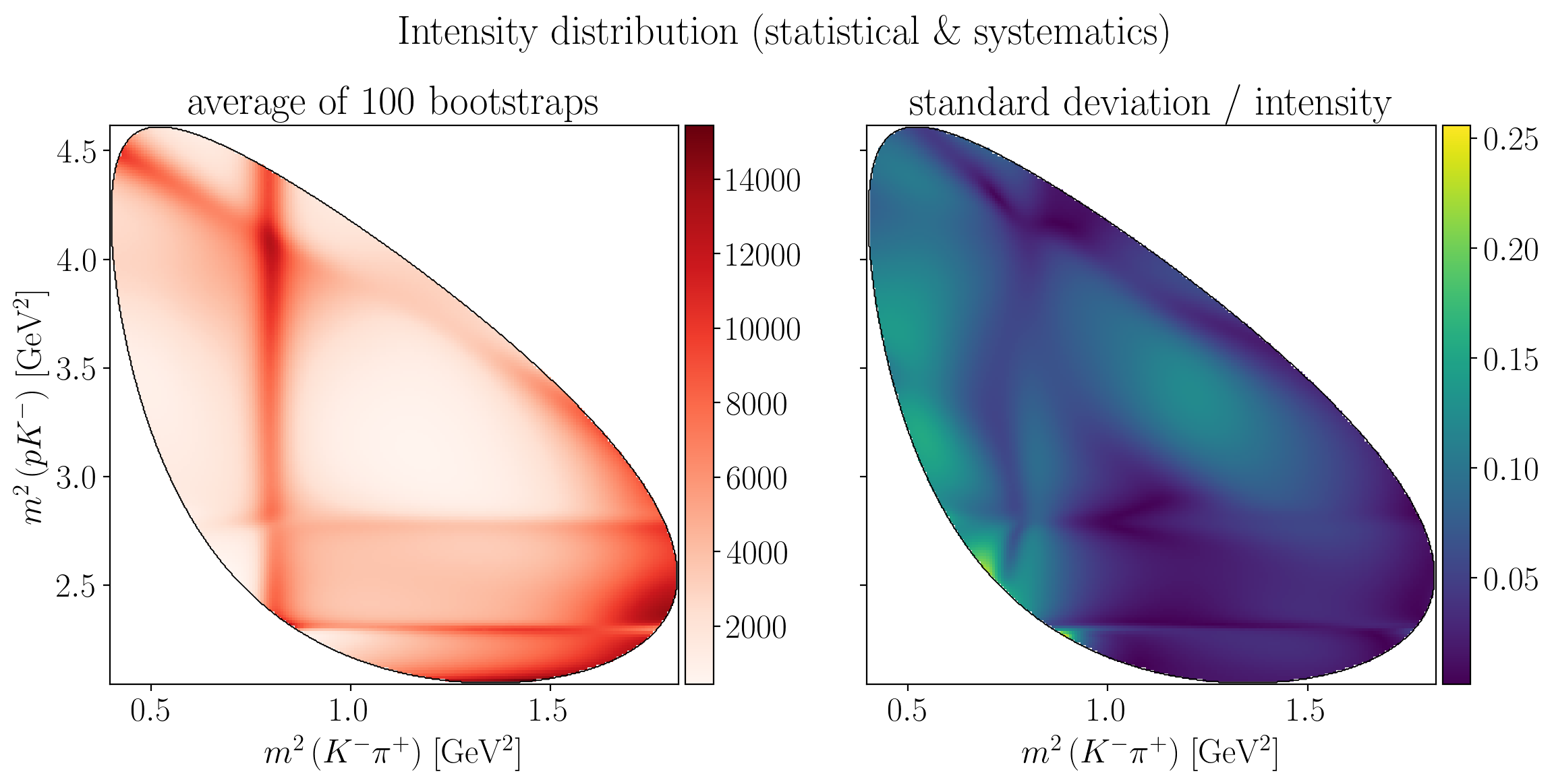
5.2.4. Comparison with nominal values#
Compute nominal values
for func in nominal_functions.values():
func.update_parameters(nominal_parameters)
nominal_intensity = nominal_functions["intensity"](phsp_sample)
nominal_polarimetry = jnp.array(
[nominal_functions[f"alpha_{i}"](phsp_sample).real for i in "xyz"]
)
nominal_polarimetry_norm = jnp.sqrt(jnp.sum(nominal_polarimetry**2, axis=0))
Show code cell source
%config InlineBackend.figure_formats = ['png']
fig, axes = plt.subplots(
dpi=200,
figsize=(17.3, 4),
gridspec_kw={"width_ratios": [1, 1, 1, 1.2]},
ncols=4,
sharey=True,
)
plt.subplots_adjust(hspace=0.2, wspace=0.05)
fig.suptitle("Comparison with nominal values", y=1.04)
axes[0].set_ylabel(s2_label)
for ax in axes:
ax.set_xlabel(s1_label)
vmax = 5.0 # %
for i in range(4):
if i != 0:
title = Rf"$\alpha_{'xyz'[i-1]}$"
z_values = jnp.abs(
(stat_alpha_mean[i] - nominal_polarimetry[i - 1])
/ nominal_polarimetry[i - 1]
)
else:
title = "$I$"
z_values = 100 * jnp.abs(
(stat_intensity_mean - nominal_intensity) / nominal_intensity
)
axes[i].set_title(title)
draw_dalitz_contour(axes[i], zorder=10)
Z = interpolate_to_grid(z_values)
mesh = axes[i].pcolormesh(X, Y, Z, cmap=cm.Reds)
mesh.set_clim(vmin=0, vmax=vmax)
if axes[i] is axes[-1]:
c_bar = fig.colorbar(mesh, ax=axes[i], pad=0.02)
c_bar.ax.set_ylabel(R"difference with nominal value (\%)")
plt.show()
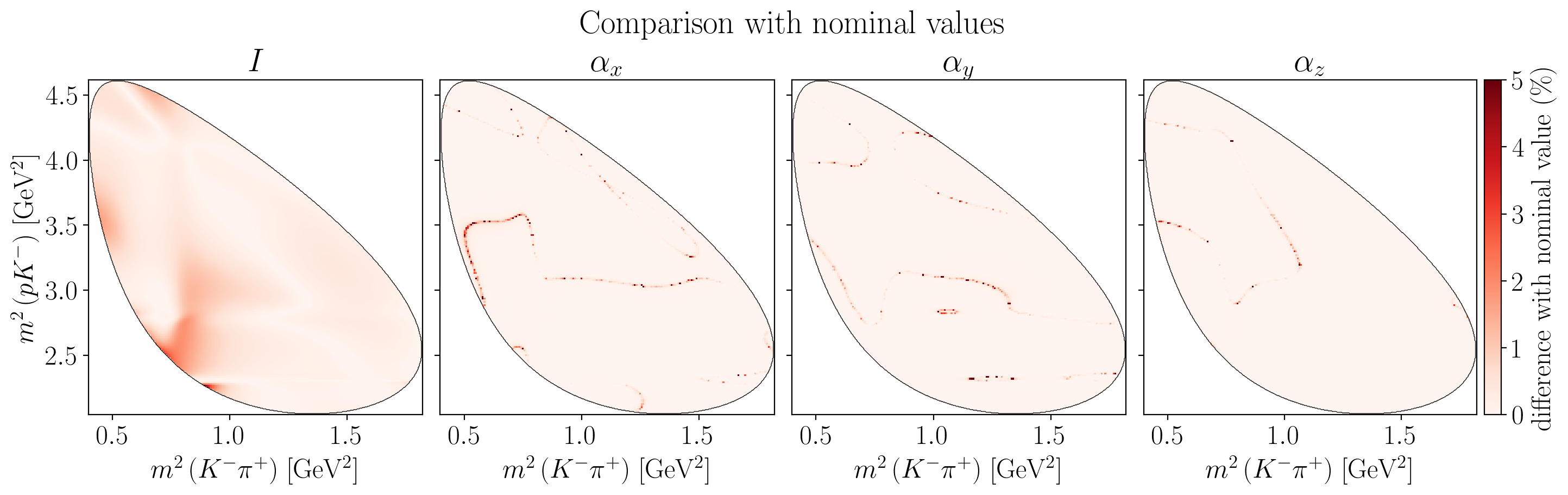
5.3. Systematic uncertainties#
Show code cell content
syst_grids = defaultdict(list)
syst_decay_rates = defaultdict(list)
progress_bar = tqdm(desc="Computing systematics", disable=NO_TQDM, total=len(models))
for title, model in models.items():
progress_bar.set_postfix_str(title)
for key, func in jax_functions[title].items():
func.update_parameters(original_parameters[title])
syst_grids[key].append(func(phsp_sample).real)
I_tot = integrate_intensity(syst_grids["intensity"][-1])
intensity_func = jax_functions[title]["intensity"]
for resonance in resonances:
res_filter = resonance.name.replace("(", R"\(").replace(")", R"\)")
I_sub = sub_intensity(intensity_func, phsp_sample, [res_filter])
syst_decay_rates[resonance.name].append(I_sub / I_tot)
progress_bar.update()
progress_bar.set_postfix_str("")
progress_bar.close()
syst_intensities = jnp.array(syst_grids["intensity"])
syst_polarimetry = jnp.array([syst_grids[f"alpha_{i}"] for i in "xyz"])
syst_polarimetry_norm = jnp.sqrt(jnp.sum(syst_polarimetry**2, axis=0))
syst_decay_rates = {k: jnp.array(v) for k, v in syst_decay_rates.items()}
5.3.1. Mean and standard deviations#
n_models = len(models)
assert syst_intensities.shape == (n_models, n_events)
assert syst_polarimetry.shape == (3, n_models, n_events)
assert syst_polarimetry_norm.shape == (n_models, n_events)
n_models, n_events
(18, 100000)
Compute statistical measures (mean, std, etc.)
syst_alpha_mean = [
jnp.mean(syst_polarimetry_norm, axis=0),
*jnp.mean(syst_polarimetry, axis=1),
]
alpha_diff_with_model_0 = [
syst_polarimetry_norm - syst_polarimetry_norm[0],
*(syst_polarimetry - syst_polarimetry[:, None, 0]),
]
syst_alpha_mean = jnp.array(syst_alpha_mean)
alpha_diff_with_model_0 = jnp.array(alpha_diff_with_model_0)
assert alpha_diff_with_model_0.shape == (4, n_models, n_events)
assert jnp.nanmax(alpha_diff_with_model_0[:, 0]) == 0.0
alpha_syst_extrema = jnp.abs(alpha_diff_with_model_0).max(axis=1)
syst_polarimetry_times_I = [
syst_polarimetry_norm * syst_intensities,
*(syst_polarimetry * syst_intensities),
]
syst_polarimetry_times_I = jnp.array(syst_polarimetry_times_I)
syst_alpha_times_I_mean = syst_polarimetry_times_I.mean(axis=1)
syst_alpha_times_I_diff = (
syst_polarimetry_times_I - syst_polarimetry_times_I[:, None, 0]
)
assert syst_alpha_times_I_diff.shape == (4, n_models, n_events)
assert jnp.nanmax(syst_alpha_times_I_diff[:, 0]) == 0.0
syst_alpha_times_I_extrema = jnp.abs(syst_alpha_times_I_diff).max(axis=1)
intensity_diff_with_model_0 = syst_intensities - syst_intensities[0]
intensity_extrema = jnp.nanmax(intensity_diff_with_model_0, axis=0)
5.3.2. Distributions#
Show 1D projections of each model
def plot_intensity_distributions(sigma: int) -> None:
original_font_size = plt.rcParams["font.size"]
use_mpl_latex_fonts()
plt.rc("font", size=10)
n_subplots = n_models - 1
nrows = int(np.floor(np.sqrt(n_subplots)))
ncols = int(np.ceil(n_subplots / nrows))
fig, axes = plt.subplots(
figsize=2.0 * np.array([ncols, nrows]),
dpi=200,
ncols=ncols,
nrows=nrows,
sharex=True,
sharey=True,
)
fig.subplots_adjust(hspace=0, wspace=0, bottom=0.1, left=0.06)
x_label = {1: s1_label, 2: s2_label, 3: s3_label}[sigma]
fig.text(0.5, 0.04, x_label, ha="center")
fig.text(0.04, 0.5, "$I$ (normalized)", va="center", rotation="vertical")
for ax in axes.flatten()[n_subplots:]:
fig.delaxes(ax)
n_bins = 80
nominal_bin_values, bin_edges = jnp.histogram(
phsp_sample[f"sigma{sigma}"],
weights=syst_intensities[0],
bins=n_bins,
density=True,
)
for i, (ax, intensities) in enumerate(
zip(axes.flatten(), syst_intensities[1:]), 1
):
ax.set_title(f"Model {i}", y=0.01)
ax.set_yticks([])
bin_values, _ = jnp.histogram(
phsp_sample[f"sigma{sigma}"],
weights=intensities,
bins=n_bins,
density=True,
)
ax.fill_between(
bin_edges[:-1],
bin_values,
alpha=0.5,
step="pre",
)
ax.step(
x=bin_edges[:-1],
y=nominal_bin_values,
linewidth=0.3,
color="red",
)
fig.savefig(f"_images/intensity-distributions-sigma{sigma}.svg")
plt.show()
use_mpl_latex_fonts()
plt.rc("font", size=original_font_size)
%config InlineBackend.figure_formats = ['svg']
plot_intensity_distributions(1)
plot_intensity_distributions(2)
plot_intensity_distributions(3)
Show code cell source
%config InlineBackend.figure_formats = ['png']
fig, axes = plt.subplots(
dpi=200,
figsize=(16.7, 8),
gridspec_kw={"width_ratios": [1, 1, 1, 1.18]},
ncols=4,
nrows=2,
sharex=True,
sharey=True,
)
plt.subplots_adjust(hspace=0.02, wspace=0.02)
fig.suptitle(R"Polarimetry field $\vec\alpha$ (model)")
axes[0, 0].set_ylabel(s2_label)
axes[1, 0].set_ylabel(s2_label)
global_max_std = jnp.nanmax(alpha_syst_extrema)
for i in range(4):
if i != 0:
title = Rf"$\alpha_{'xyz'[i-1]}$"
else:
title = R"$\left|\vec\alpha\right|$"
axes[0, i].set_title(title)
draw_dalitz_contour(axes[0, i], zorder=10)
Z = interpolate_to_grid(syst_alpha_mean[i])
mesh = axes[0, i].pcolormesh(X, Y, Z, cmap=cm.RdYlGn_r)
mesh.set_clim(vmin=-1, vmax=+1)
if axes[0, i] is axes[0, -1]:
c_bar = fig.colorbar(mesh, ax=axes[0, i], pad=0.01)
c_bar.ax.set_ylabel(Rf"$\alpha$ averaged with {n_models} models")
c_bar.ax.set_yticks([-1, 0, +1])
c_bar.ax.set_yticklabels(["-1", "0", "+1"])
draw_dalitz_contour(axes[1, i], zorder=10)
Z = interpolate_to_grid(alpha_syst_extrema[i])
mesh = axes[1, i].pcolormesh(X, Y, Z)
mesh.set_clim(vmin=0, vmax=global_max_std)
axes[1, i].set_xlabel(s1_label)
if axes[1, i] is axes[1, -1]:
c_bar = fig.colorbar(mesh, ax=axes[1, i], pad=0.01)
c_bar.ax.set_ylabel("maximum absolute difference\nwith the default model")
plt.show()
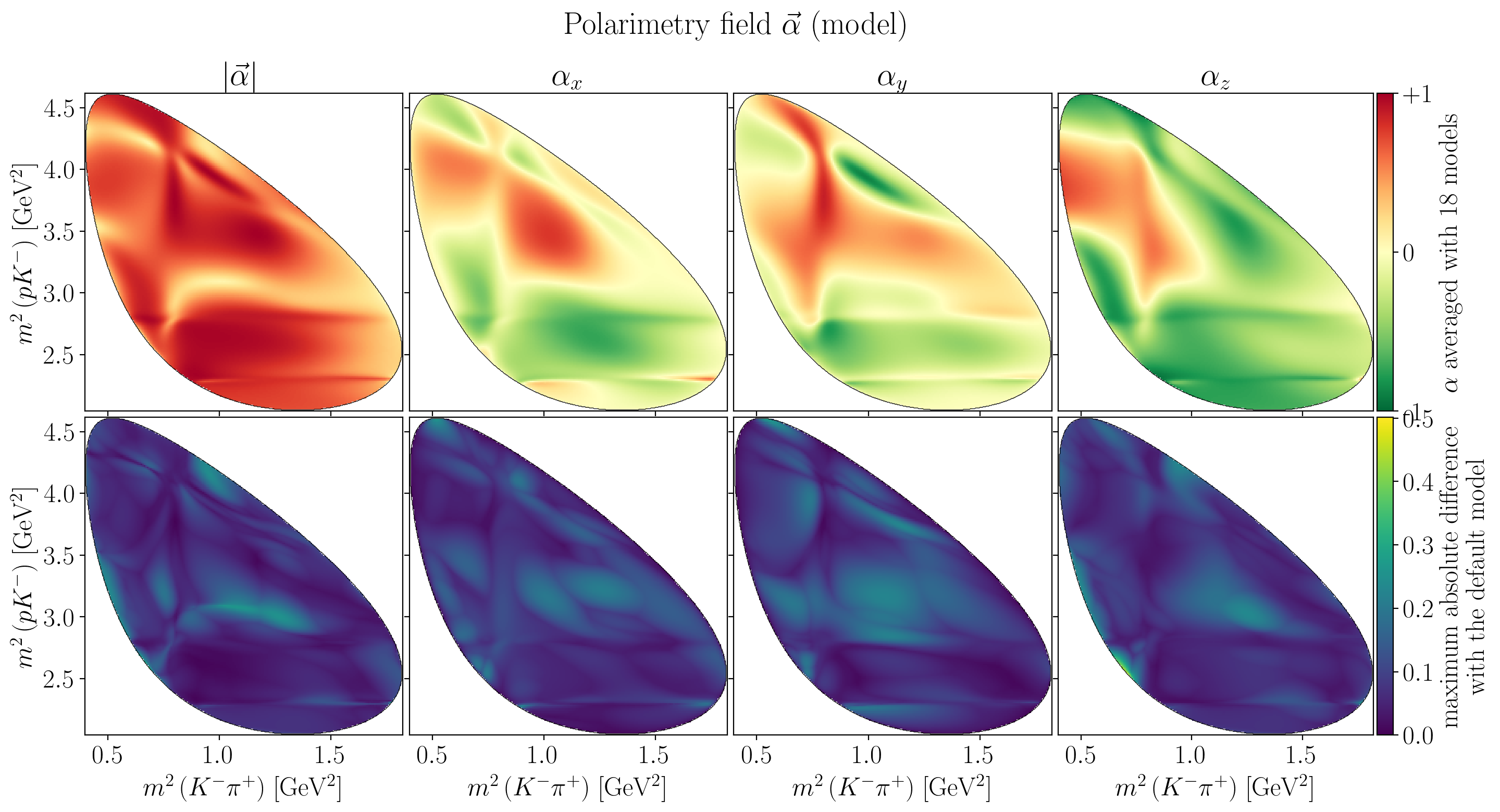
Show code cell source
%config InlineBackend.figure_formats = ['png']
fig, axes = plt.subplots(
dpi=200,
figsize=(16.7, 8),
gridspec_kw={"width_ratios": [1, 1, 1, 1.18]},
ncols=4,
nrows=2,
sharex=True,
sharey=True,
)
plt.subplots_adjust(hspace=0.02, wspace=0.02)
axes[0, 0].set_ylabel(s2_label)
axes[1, 0].set_ylabel(s2_label)
syst_intensity_mean = jnp.mean(syst_intensities, axis=0)
global_max_mean = jnp.nanmax(jnp.abs(syst_alpha_times_I_mean))
global_max_diff = jnp.nanmax(syst_alpha_times_I_extrema / syst_intensity_mean)
for i in range(4):
if i != 0:
title = Rf"$\alpha_{'xyz'[i-1]}$"
else:
title = R"$\left|\vec\alpha\right|$"
axes[0, i].set_title(title)
axes[1, i].set_xlabel(s1_label)
draw_dalitz_contour(axes[0, i], zorder=10)
Z = interpolate_to_grid(syst_alpha_times_I_mean[i])
mesh = axes[0, i].pcolormesh(X, Y, Z, cmap=cm.RdYlGn_r)
mesh.set_clim(vmin=-global_max_mean, vmax=+global_max_mean)
if axes[0, i] is axes[0, -1]:
c_bar = fig.colorbar(mesh, ax=axes[0, i], pad=0.01)
c_bar.ax.set_ylabel(Rf"$\alpha \cdot I$ averaged with {n_models} models")
draw_dalitz_contour(axes[1, i], zorder=10)
Z = interpolate_to_grid(syst_alpha_times_I_extrema[i] / syst_intensity_mean)
mesh = axes[1, i].pcolormesh(X, Y, Z)
mesh.set_clim(vmin=0, vmax=global_max_diff)
if axes[1, i] is axes[1, -1]:
c_bar = fig.colorbar(mesh, ax=axes[1, i], pad=0.01)
c_bar.ax.set_ylabel(
"maximum absolute difference\n"
"with the default model\n"
"divided by nominal intensity"
)
plt.show()
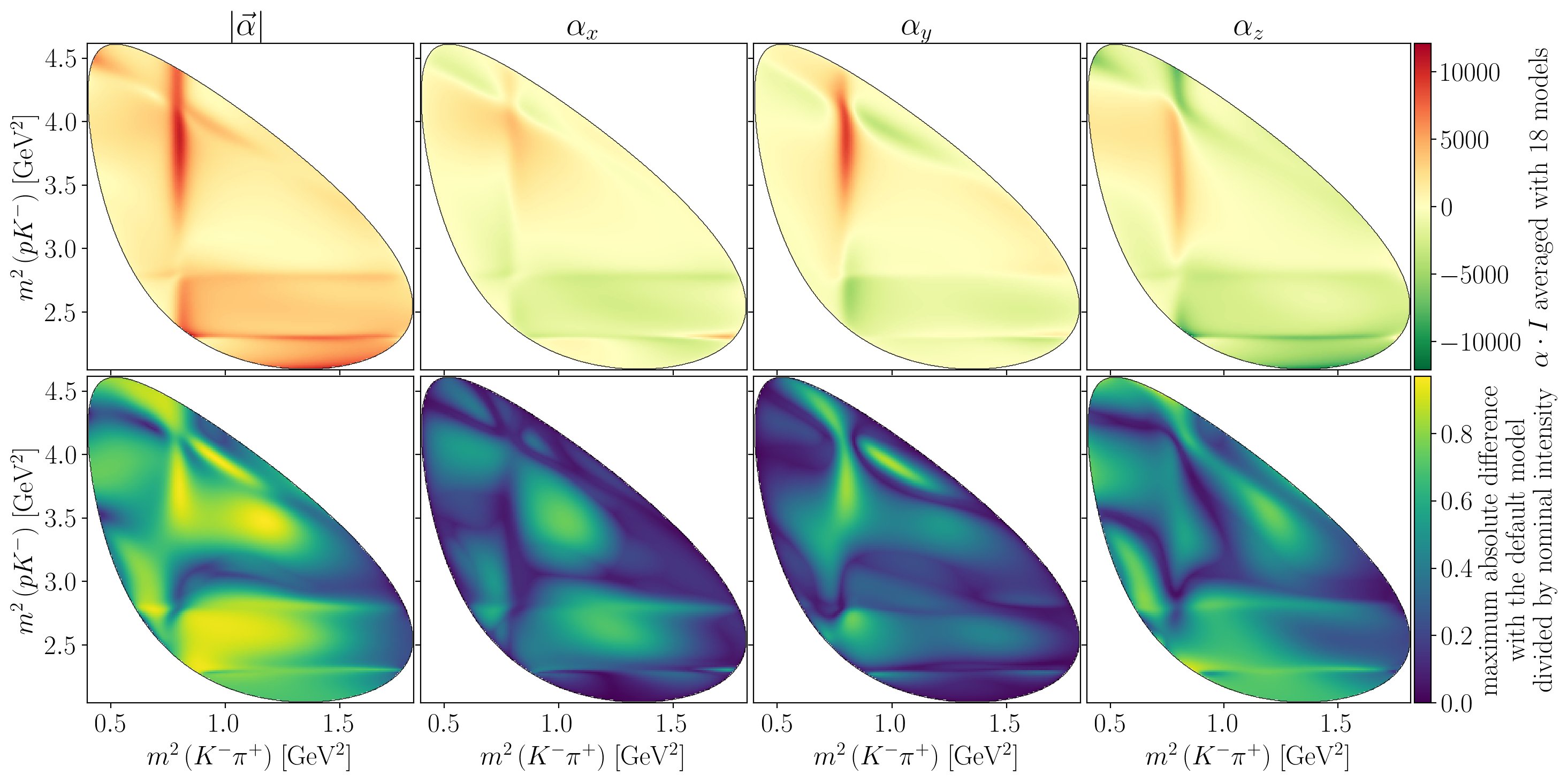
Show code cell source
%config InlineBackend.figure_formats = ['png']
fig, (ax1, ax2) = plt.subplots(
dpi=200,
figsize=(12, 6.9),
ncols=2,
sharey=True,
)
fig.suptitle("Intensity distribution (model)", y=0.95)
ax1.set_xlabel(s1_label)
ax2.set_xlabel(s1_label)
ax1.set_ylabel(s2_label)
Z = interpolate_to_grid(syst_intensity_mean)
mesh = ax1.pcolormesh(X, Y, Z, cmap=cm.Reds)
fig.colorbar(mesh, ax=ax1, pad=0.01)
draw_dalitz_contour(ax1, width=0.2)
ax1.set_title(f"average of {n_models} models")
Z = interpolate_to_grid(intensity_extrema / syst_intensity_mean)
mesh = ax2.pcolormesh(X, Y, Z)
fig.colorbar(mesh, ax=ax2, pad=0.01)
draw_dalitz_contour(ax2, width=0.2)
ax2.set_title("maximum absolute difference\n" R"with the default model (\%)")
fig.tight_layout()
plt.show()
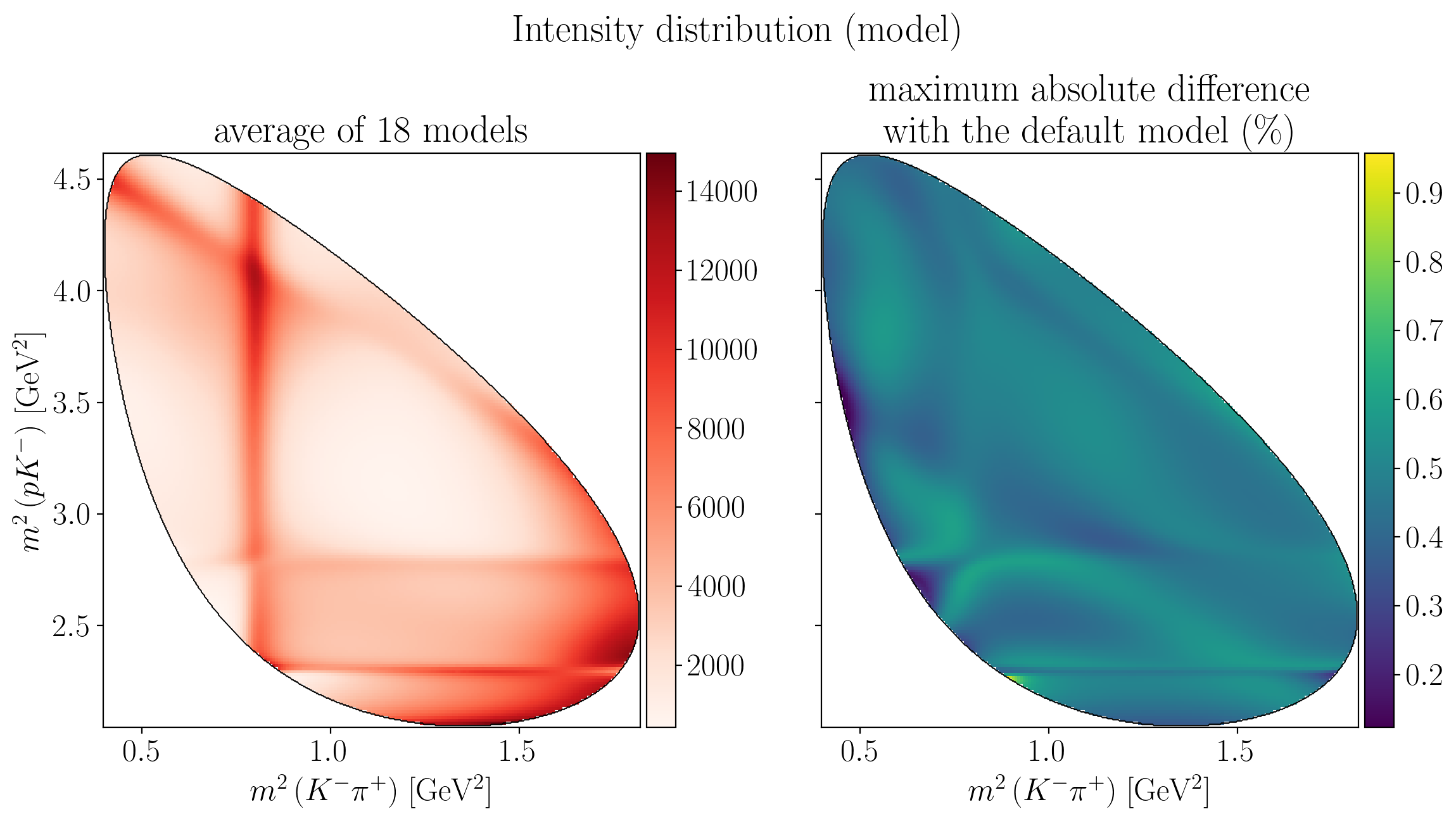
5.4. Uncertainty on polarimetry#
For each bootstrap or alternative model \(i\), we compute the angle between each aligned polarimeter vector \(\vec\alpha_i\) and the one from the nominal model, \(\vec\alpha_0\):
The solid angle can then be computed as:
The statistical uncertainty is given by taking the standard deviation on the \(\delta\Omega\) distribution and the systematic uncertainty is given by taking finding \(\theta_\mathrm{max} = \max\theta_i\) and computing \(\delta\Omega_\mathrm{max}\) from that.
Show code cell source
def dot(array1: jnp.ndarray, array2: jnp.ndarray, axis=0) -> jnp.ndarray:
return jnp.sum(array1 * array2, axis=axis)
def norm(array: jnp.ndarray, axis=0) -> jnp.ndarray:
return jnp.sqrt(dot(array, array, axis=axis))
def compute_theta(polarimetry: jnp.ndarray) -> jnp.ndarray:
return jnp.arccos(
dot(polarimetry, nominal_polarimetry[:, None])
/ (norm(polarimetry) * norm(nominal_polarimetry))
)
def compute_solid_angle(theta: jnp.ndarray) -> jnp.ndarray:
return 2 * jnp.pi * (1 - jnp.cos(theta))
stat_theta = compute_theta(stat_polarimetry)
syst_theta = compute_theta(syst_polarimetry)
assert stat_theta.shape == (n_bootstraps, n_events)
assert syst_theta.shape == (n_models, n_events)
stat_solid_angle = jnp.nanstd(compute_solid_angle(stat_theta), axis=0)
syst_solid_angle = jnp.nanmax(compute_solid_angle(syst_theta), axis=0)
def plot_angle_uncertainties(watermark: bool, titles: bool) -> None:
plt.ioff()
fig, axes = plt.subplots(
dpi=200,
figsize=(11, 4),
ncols=2,
)
fig.subplots_adjust(wspace=0.3)
ax1, ax2 = axes
if titles:
fig.suptitle(R"Uncertainty over $\vec\alpha$ polar angle", y=1.04)
ax1.set_title(R"Statistical \& systematic")
ax2.set_title("Model")
for ax in axes:
ax.set_box_aspect(1)
ax.set_xlabel(s1_label)
ax.set_ylabel(s2_label)
draw_dalitz_contour(ax, width=0.2, zorder=10)
Z = interpolate_to_grid(stat_solid_angle)
mesh = ax1.pcolormesh(X, Y, Z, cmap=plt.cm.YlOrRd)
mesh.set_clim(0, 4 * np.pi)
c_bar = fig.colorbar(mesh, ax=ax1, pad=0.02)
c_bar.ax.set_ylabel(R"Std. of $\delta\Omega$")
c_bar.ax.set_yticks([0, 2 * np.pi, 4 * np.pi])
c_bar.ax.set_yticklabels(["$0$", R"$2\pi$", R"$4\pi$"])
Z = interpolate_to_grid(syst_solid_angle)
mesh = ax2.pcolormesh(X, Y, Z, cmap=plt.cm.YlOrRd)
mesh.set_clim(0, 4 * np.pi)
c_bar = fig.colorbar(mesh, ax=ax2, pad=0.02)
c_bar.ax.set_ylabel(R"Max. of $\delta\Omega$")
c_bar.ax.set_yticks([0, 2 * np.pi, 4 * np.pi])
c_bar.ax.set_yticklabels(["$0$", R"$2\pi$", R"$4\pi$"])
output_file = "polarimetry-field-angle-uncertainties"
if watermark:
output_file += "-watermark"
add_watermark_to_uncertainties(ax1)
add_watermark_to_uncertainties(ax2)
if titles:
output_file += "-with-titles"
fig.savefig(f"_static/images/{output_file}.png", bbox_inches="tight")
if watermark and titles:
plt.show()
plt.close(fig)
plt.ion()
def add_watermark_to_uncertainties(ax) -> None:
add_watermark(ax, 0.70, 0.82, fontsize=16)
%config InlineBackend.figure_formats = ['png']
plt.rcdefaults()
use_mpl_latex_fonts()
plt.rc("font", size=16)
boolean_combinations = list(itertools.product(*2 * [[False, True]]))
for watermark, titles in tqdm(boolean_combinations, disable=NO_TQDM):
plot_angle_uncertainties(watermark, titles)
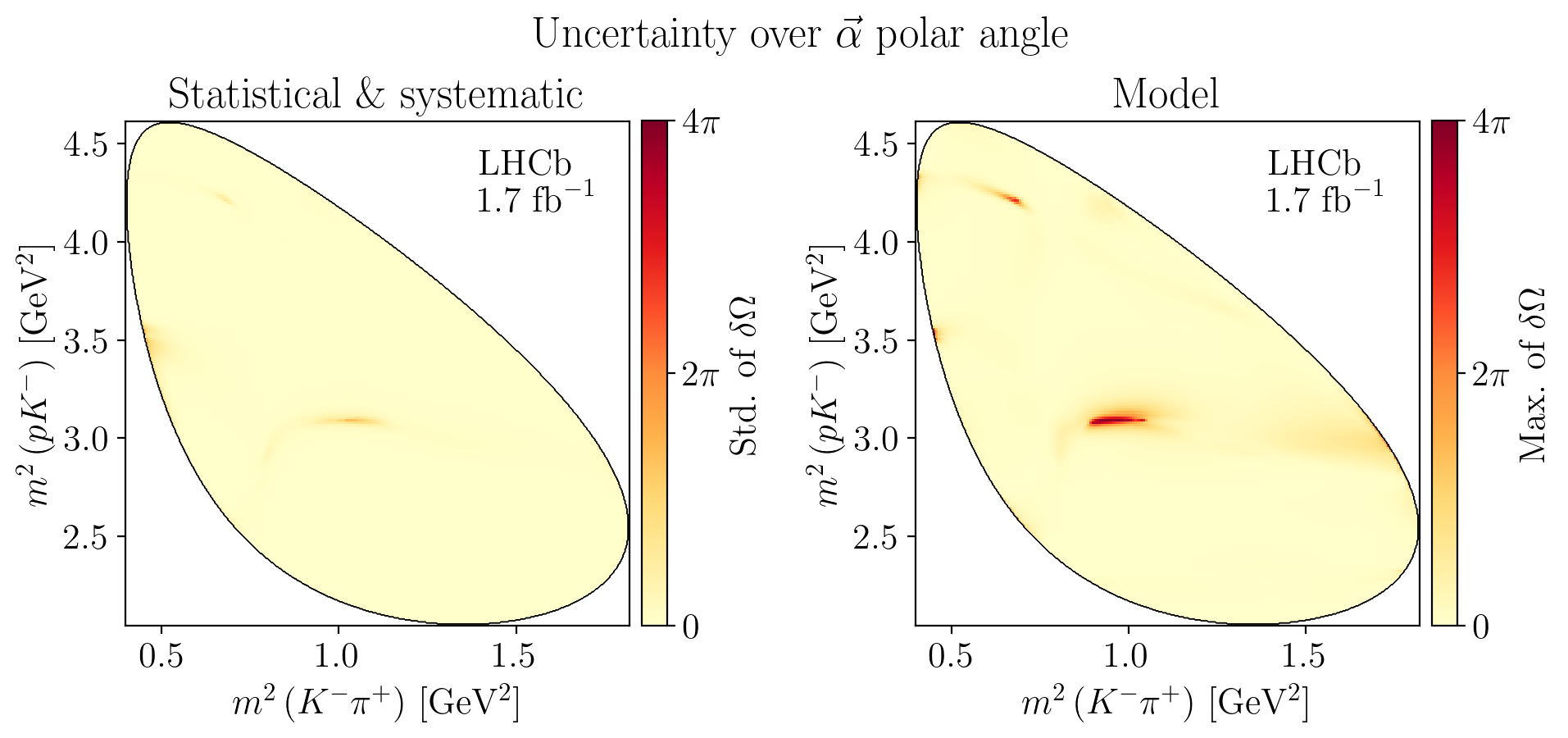
Show code cell source
stat_alpha_difference = norm(stat_polarimetry - nominal_polarimetry[:, None])
syst_alpha_difference = norm(syst_polarimetry - nominal_polarimetry[:, None])
nominal_polarimetry_norm = norm(nominal_polarimetry[:, None])
stat_alpha_rel_difference = 100 * stat_alpha_difference / nominal_polarimetry_norm
syst_alpha_rel_difference = 100 * syst_alpha_difference / nominal_polarimetry_norm
assert stat_alpha_rel_difference.shape == (n_bootstraps, n_events)
assert syst_alpha_rel_difference.shape == (n_models, n_events)
def create_figure(titles: bool):
fig, axes = plt.subplots(
dpi=200,
figsize=(11.5, 4),
ncols=2,
)
fig.subplots_adjust(wspace=0.4)
ax1, ax2 = axes
if titles:
fig.suptitle(R"Uncertainty over $\left|\vec\alpha\right|$", y=1.04)
ax1.set_title(R"Statistical \& systematic")
ax2.set_title("Model")
for ax in axes:
ax.set_box_aspect(1)
ax.set_xlabel(s1_label)
ax.set_ylabel(s2_label)
draw_dalitz_contour(ax, width=0.2)
return fig, (ax1, ax2)
def plot_norm_rel_uncertainties(watermark: bool, titles: bool) -> None:
plt.ioff()
fig, (ax1, ax2) = create_figure(titles)
Z = interpolate_to_grid(jnp.nanstd(stat_alpha_rel_difference, axis=0))
mesh = ax1.pcolormesh(X, Y, Z, cmap=plt.cm.YlOrRd)
mesh.set_clim(0, 100)
c_bar = fig.colorbar(mesh, ax=ax1, pad=0.02)
c_bar.ax.set_ylabel(
R"Std. of"
R" $\frac{|\alpha^{(i)}-\alpha^\mathrm{default}|}{|\alpha^\mathrm{default}|}$"
)
c_bar.ax.set_yticks([0, 50, 100])
c_bar.ax.set_yticklabels([R"0\%", R"50\%", R"100\%"])
Z = interpolate_to_grid(jnp.nanmax(syst_alpha_rel_difference, axis=0))
mesh = ax2.pcolormesh(X, Y, Z, cmap=plt.cm.YlOrRd)
mesh.set_clim(0, 100)
c_bar = fig.colorbar(mesh, ax=ax2, pad=0.02)
c_bar.ax.set_ylabel(
R"Max. of"
R" $\frac{|\alpha^{(i)}-\alpha^\mathrm{default}|}{|\alpha^\mathrm{default}|}$"
)
c_bar.ax.set_yticks([0, 50, 100])
c_bar.ax.set_yticklabels([R"0\%", R"50\%", R"100\%"])
output_file = "polarimetry-field-norm-uncertainties-relative"
if watermark:
output_file += "-watermark"
add_watermark_to_uncertainties(ax1)
add_watermark_to_uncertainties(ax2)
if titles:
output_file += "-with-titles"
fig.savefig(f"_static/images/{output_file}.png", bbox_inches="tight")
if watermark and titles:
plt.show()
plt.close(fig)
plt.ion()
def plot_norm_uncertainties(watermark: bool, titles: bool) -> None:
plt.ioff()
vmax = 0.5
fig, (ax1, ax2) = create_figure(titles)
Z = interpolate_to_grid(jnp.nanstd(stat_alpha_difference, axis=0))
mesh = ax1.pcolormesh(X, Y, Z, cmap=plt.cm.YlOrRd)
c_bar = fig.colorbar(mesh, ax=ax1, pad=0.02)
mesh.set_clim(0, vmax)
c_bar.ax.set_ylabel(R"Std. of $|\alpha^{(i)}-\alpha^\mathrm{default}|$")
Z = interpolate_to_grid(jnp.nanmax(syst_alpha_difference, axis=0))
mesh = ax2.pcolormesh(X, Y, Z, cmap=plt.cm.YlOrRd)
mesh.set_clim(0, vmax)
c_bar = fig.colorbar(mesh, ax=ax2, pad=0.02)
c_bar.ax.set_ylabel(R"Max. of $|\alpha^{(i)}-\alpha^\mathrm{default}|$")
output_file = "polarimetry-field-norm-uncertainties"
if watermark:
output_file += "-watermark"
add_watermark_to_uncertainties(ax1)
add_watermark_to_uncertainties(ax2)
if titles:
output_file += "-with-titles"
fig.savefig(f"_static/images/{output_file}.png", bbox_inches="tight")
if watermark and titles:
plt.show()
plt.close(fig)
plt.ion()
%config InlineBackend.figure_formats = ['png']
plt.rcdefaults()
use_mpl_latex_fonts()
plt.rc("font", size=18)
for watermark, titles in tqdm(boolean_combinations, disable=NO_TQDM):
plot_norm_rel_uncertainties(watermark, titles)
plot_norm_uncertainties(watermark, titles)
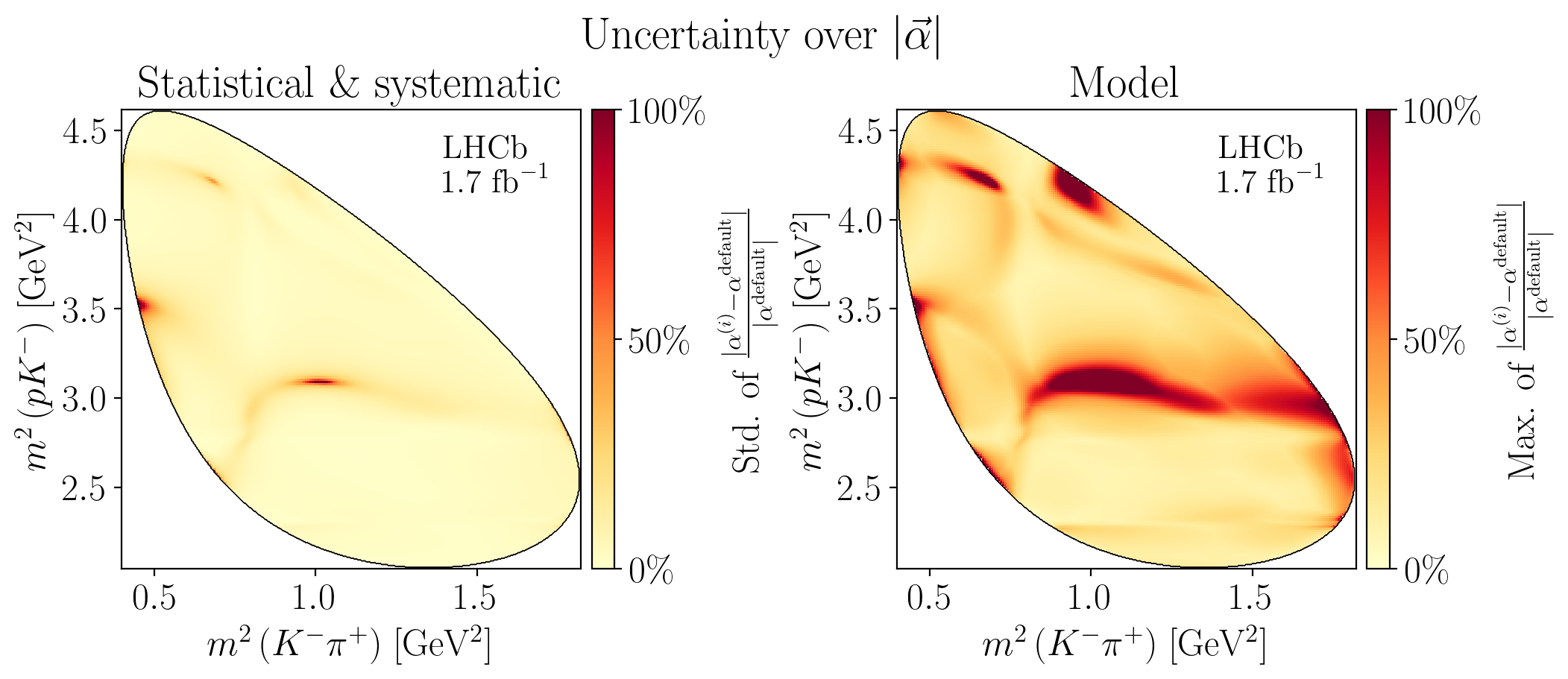
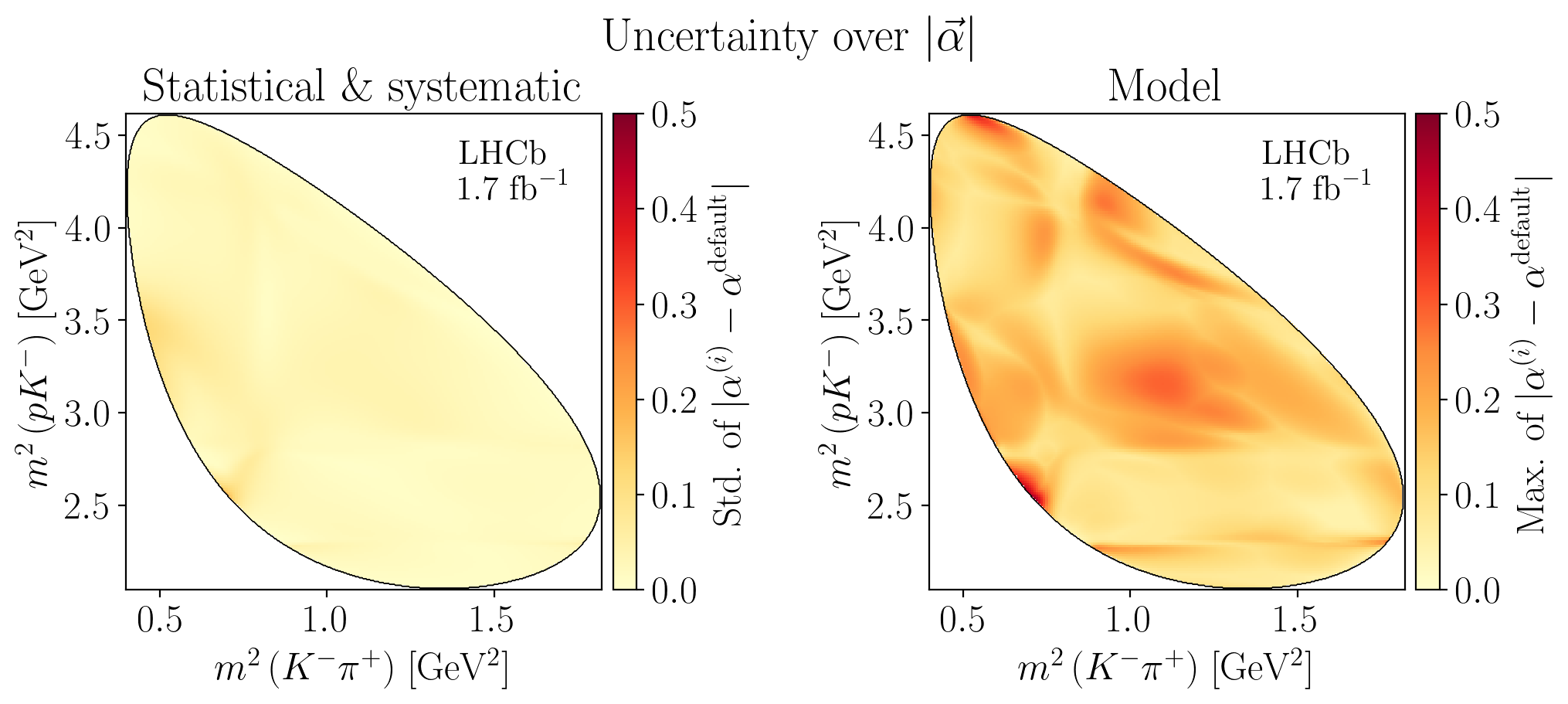
5.5. Decay rates#
Show code cell source
with open("../data/observable-references.yaml") as f:
data = yaml.load(f, Loader=yaml.SafeLoader)
lhcb_values = {
k: tuple(float(v) for v in row.split(" ± "))
for k, row in data[nominal_model_title]["rate"].items()
}
src = R"""
\begin{array}{l|c|c}
\textbf{Resonance} & \textbf{Decay rate} & \textbf{LHCb} \\
\hline
"""
for resonance in resonances:
ff_statistical = 100 * stat_decay_rates[resonance.name]
ff_systematics = 100 * syst_decay_rates[resonance.name]
ff_nominal = f"{ff_systematics[0]:.2f}"
ff_stat = f"{ff_statistical.std():.2f}"
ff_syst_min = f"{(ff_systematics[1:]-ff_systematics[0]).min():+.2f}"
ff_syst_max = f"{(ff_systematics[1:]-ff_systematics[0]).max():+.2f}"
src += f"{resonance.latex} & "
src += Rf"{ff_nominal} \pm {ff_stat}_{{{ff_syst_min}}}^{{{ff_syst_max}}} & "
lhcb_value, lhcb_stat, lhcb_syst, _ = lhcb_values[resonance.name]
src += Rf"{lhcb_value} \pm {lhcb_stat} \pm {lhcb_syst} \\" "\n"
src += R"\end{array}"
Latex(src)
Show code cell source
alignment = "c".join("" for _ in range(n_models) if i)
names = " & ".join(Rf"\textbf{{{i}}}" for i in range(n_models) if i)
src = Rf"""
$$
\begin{{array}}{{l|{alignment}}}
\textbf{{Resonance}} & {names} \\
\hline
"""
for resonance in resonances:
ff_systematics = 100 * syst_decay_rates[resonance.name]
src += f" {resonance.latex:13s}"
for ff_model in ff_systematics[1:]:
diff = f"{ff_model-ff_systematics[0]:+.2f}"
if ff_model == ff_systematics[1:].min():
src += Rf" & \color{{blue}}{{{diff}}}"
elif ff_model == ff_systematics[1:].max():
src += Rf" & \color{{red}}{{{diff}}}"
else:
src += f" & {diff}"
src += R" \\" "\n"
src += """\
\\end{array}
$$
"""
for i, title in enumerate(models):
src += f"\n- **{i}**: {title}"
Markdown(src)
0: Default amplitude model
1: Alternative amplitude model with K(892) with free mass and width
2: Alternative amplitude model with L(1670) with free mass and width
3: Alternative amplitude model with L(1690) with free mass and width
4: Alternative amplitude model with D(1232) with free mass and width
5: Alternative amplitude model with L(1600), D(1600), D(1700) with free mass and width
6: Alternative amplitude model with free L(1405) Flatt’e widths, indicated as G1 (pK channel) and G2 (Sigmapi)
7: Alternative amplitude model with L(1800) contribution added with free mass and width
8: Alternative amplitude model with L(1810) contribution added with free mass and width
9: Alternative amplitude model with D(1620) contribution added with free mass and width
10: Alternative amplitude model in which a Relativistic Breit-Wigner is used for the K(700) contribution
11: Alternative amplitude model with K(700) with free mass and width
12: Alternative amplitude model with K(1410) contribution added with mass and width from PDG2020
13: Alternative amplitude model in which a Relativistic Breit-Wigner is used for the K(1430) contribution
14: Alternative amplitude model with K(1430) with free width
15: Alternative amplitude model with an additional overall exponential form factor exp(-alpha q^2) multiplying Bugg lineshapes. The exponential parameter is indicated as ``alpha’’
16: Alternative amplitude model with free radial parameter d for the Lc resonance, indicated as dLc
17: Alternative amplitude model obtained using LS couplings
5.6. Average polarimetry values#
The components of the averaged polarimeter vector \(\overline{\alpha}\) are defined as:
The averages of the norm of \(\vec\alpha\) are computed as follows:
\(\left|\overline{\alpha}\right| = \sqrt{\overline{\alpha_x}^2+\overline{\alpha_y}^2+\overline{\alpha_z}^2}\), with the statistical uncertainties added in quadrature and the systematic uncertainties by taking the same formula on the extrema values of each \(\overline{\alpha_j}\)
\(\overline{\left|\alpha\right|} = \sqrt{\int I_0\left(\tau\right) \left|\vec\alpha\left(\tau\right)\right|^2 \mathrm{d}^n \tau \; \big / \int I_0\left(\tau\right)\,\mathrm{d}^n \tau}\)
Show code cell source
def compute_weighted_average(v: jnp.ndarray, weights: jnp.ndarray) -> jnp.ndarray:
return jnp.nansum(v * weights, axis=-1) / jnp.nansum(weights, axis=-1)
def compute_polar_coordinates(cartesian_vector: jnp.ndarray) -> jnp.ndarray:
x, y, z = cartesian_vector
norm = jnp.sqrt(jnp.sum(cartesian_vector**2, axis=0))
theta = np.arccos(z / norm)
phi = np.pi - np.arctan2(y, -x)
return jnp.array([norm, theta, phi])
stat_weighted_alpha_norm = jnp.sqrt(
compute_weighted_average(stat_polarimetry_norm**2, weights=stat_intensities)
)
stat_weighted_alpha = compute_weighted_average(
stat_polarimetry,
weights=stat_intensities,
)
stat_weighted_alpha_polar = compute_polar_coordinates(stat_weighted_alpha)
assert stat_weighted_alpha_norm.shape == (n_bootstraps,)
assert stat_weighted_alpha.shape == (3, n_bootstraps)
assert stat_weighted_alpha_polar.shape == (3, n_bootstraps)
syst_weighted_alpha_norm = jnp.sqrt(
compute_weighted_average(syst_polarimetry_norm**2, weights=syst_intensities)
)
syst_weighted_alpha = compute_weighted_average(
syst_polarimetry,
weights=syst_intensities,
)
syst_weighted_alpha_polar = compute_polar_coordinates(syst_weighted_alpha)
assert syst_weighted_alpha_norm.shape == (n_models,)
assert syst_weighted_alpha.shape == (3, n_models)
assert syst_weighted_alpha_polar.shape == (3, n_models)
nominal_weighted_alpha_norm = syst_weighted_alpha_norm[0]
nominal_weighted_alpha = syst_weighted_alpha[:, 0]
nominal_weighted_alpha_polar = syst_weighted_alpha_polar[:, 0]
syst_weighted_alpha_norm_diff = (
syst_weighted_alpha_norm - nominal_weighted_alpha_norm
)
syst_weighted_alpha_diff = (syst_weighted_alpha.T - nominal_weighted_alpha).T
syst_weighted_alpha_diff_polar = (
syst_weighted_alpha_polar.T - nominal_weighted_alpha_polar
).T
stat_weighted_alpha_std = stat_weighted_alpha.std(axis=1)
syst_weighted_alpha_min = syst_weighted_alpha_diff.min(axis=1)
syst_weighted_alpha_max = syst_weighted_alpha_diff.max(axis=1)
stat_weighted_alpha_polar_std = stat_weighted_alpha_polar.std(axis=1)
syst_weighted_alpha_polar_min = syst_weighted_alpha_diff_polar.min(axis=1)
syst_weighted_alpha_polar_max = syst_weighted_alpha_diff_polar.max(axis=1)
Cartesian coordinates:
Show code cell source
def render_cartesian_uncertainties(
value, stat, syst_min, syst_max, plus: bool = True
) -> str:
value *= 1e3
stat *= 1e3
syst_min *= 1e3
syst_max *= 1e3
if plus:
val = f"{value:+.1f}"
else:
val = f"{value:.1f}"
stat = f"{stat:.1f}"
syst_min = f"-{abs(syst_min):.1f}"
syst_max = f"+{abs(syst_max):.1f}"
return (
Rf"\left({val} \pm {stat}_{{{syst_min}}}^{{{syst_max}}} \right) \times"
R" 10^{-3}"
)
src = R"""
\begin{array}{ccr}
"""
for i in range(3):
value = render_cartesian_uncertainties(
nominal_weighted_alpha[i],
stat_weighted_alpha_std[i],
syst_weighted_alpha_min[i],
syst_weighted_alpha_max[i],
)
src += Rf" \overline{{\alpha_{'xyz'[i]}}} & = & {value} \\" "\n"
value = render_cartesian_uncertainties(
nominal_weighted_alpha_norm,
stat_weighted_alpha_norm.std(),
syst_weighted_alpha_norm_diff.min(),
syst_weighted_alpha_norm_diff.max(),
plus=False,
)
src += Rf" \overline{{\left|\alpha\right|}} & = & {value} \\"
src += R"\end{array}"
Latex(src)
Polar coordinates:
Show code cell source
def render_radian_angle_uncertainties(value, stat, syst_min, syst_max) -> str:
val = f"{value:+.2f}"
stat = f"{stat:.2f}"
syst_min = f"-{abs(syst_min):.2f}"
syst_max = f"+{abs(syst_max):.2f}"
return Rf"{val} \pm {stat}_{{{syst_min}}}^{{{syst_max}}}\;\mathrm{{rad}}"
def render_angle_uncertainties(value, stat, syst_min, syst_max) -> str:
value /= np.pi
stat /= np.pi
syst_min /= np.pi
syst_max /= np.pi
val = f"{value:+.3f}"
stat = f"{stat:.3f}"
syst_min = f"-{abs(syst_min):.3f}"
syst_max = f"+{abs(syst_max):.3f}"
return (
Rf"\left({val} \pm {stat}_{{{syst_min}}}^{{{syst_max}}} \right) \times \pi"
)
src = R"""
\begin{array}{ccl}
"""
labels = [
R"\left|\overline{\alpha}\right|",
R"\theta\left(\overline{\alpha}\right)",
R"\phi\left(\overline{\alpha}\right)",
]
for i, label in enumerate(labels):
renderer = (
render_cartesian_uncertainties
if i == 0
else render_radian_angle_uncertainties
)
args = (
nominal_weighted_alpha_polar[i],
stat_weighted_alpha_polar_std[i],
syst_weighted_alpha_polar_min[i],
syst_weighted_alpha_polar_max[i],
)
src += Rf" {label} & = & {renderer(*args)} \\" "\n"
if i > 0:
src += Rf" & = & {render_angle_uncertainties(*args)} \\" "\n"
src += R"\end{array}"
Latex(src)
Averaged polarimeter values for each model (and the difference with the nominal model):
Show code cell source
src = R"""
\begin{array}{r|rrrr|rrrr}
\textbf{Model}
& \overline{\alpha}_x & \overline{\alpha}_y & \overline{\alpha}_z & \overline{\left|\alpha\right|}
& \Delta\overline{\alpha}_x & \Delta\overline{\alpha}_y & \Delta\overline{\alpha}_z
& \Delta\overline{\left|\alpha\right|} \\
\hline
"""
for i, title in enumerate(models):
α = 1e3 * syst_weighted_alpha[:, i]
abs_α = 1e3 * syst_weighted_alpha_norm[i]
Δα = 1e3 * syst_weighted_alpha_diff[:, i]
abs_Δα = 1e3 * syst_weighted_alpha_norm_diff[i]
src += Rf" \textbf{{{i}}}"
src += Rf" & {α[0]:+.1f} & {α[1]:+.1f} & {α[2]:+.1f} & {abs_α:.1f}"
if i != 0:
src += Rf" & {Δα[0]:+.1f} & {Δα[1]:+.1f} & {Δα[2]:+.1f} & {abs_Δα:+.1f}"
src += R" \\" "\n"
del i, title, α, abs_α, Δα, abs_Δα
src += R"\end{array}"
Latex(src)
Show code cell source
src = R"""
\begin{array}{r|rrr|rrr}
\textbf{Model}
& 10^3 \cdot \left|\overline{\alpha}\right|
& \theta\left(\overline{\alpha}\right) / \pi
& \phi\left(\overline{\alpha}\right) / \pi
& 10^3 \cdot \Delta\left|\overline{\alpha}\right|
& \Delta\theta\left(\overline{\alpha}\right) / \pi
& \Delta\phi\left(\overline{\alpha}\right) / \pi \\
\hline
"""
for i, title in enumerate(models):
α, θ, φ = syst_weighted_alpha_polar[:, i]
Δα, Δθ, Δφ = syst_weighted_alpha_diff_polar[:, i]
src += Rf" \textbf{{{i}}}"
src += Rf" & {1e3*α:+.1f} & {θ/np.pi:+.3f} & {φ/np.pi:+.3f}"
if i != 0:
src += Rf" & {1e3*Δα:+.1f} & {Δθ/np.pi:+.3f} & {Δφ/np.pi:+.3f}"
src += R" \\" "\n"
del i, title, α, θ, φ, Δα, Δθ, Δφ
src += R"\end{array}"
Latex(src)
Tip
These values can be downloaded in serialized JSON format under Exported distributions.
Show code cell source
alpha_x_reversed = syst_weighted_alpha[0, ::-1]
alpha_y_reversed = syst_weighted_alpha[1, ::-1]
alpha_z_reversed = syst_weighted_alpha[2, ::-1]
alpha_norm_reversed = syst_weighted_alpha_polar[0, ::-1]
alpha_theta_reversed = syst_weighted_alpha_polar[1, ::-1]
alpha_phi_reversed = syst_weighted_alpha_polar[2, ::-1]
marker_options = dict(
color=(n_models - 1) * ["blue"] + ["red"],
size=(n_models - 1) * [7] + [10],
opacity=0.7,
)
model_titles_reversed = [
"<br>".join(wrap(title, width=60)) for title in reversed(models)
]
fig = make_subplots(cols=2, shared_yaxes=True)
fig.add_trace(
go.Scatter(
x=alpha_phi_reversed,
y=alpha_theta_reversed,
hovertemplate="<b>%{text}</b><br>"
+ "ϕ(ɑ̅) = %{x:.3f}, "
+ "θ(ɑ̅) = %{y:.3f}"
+ "<extra></extra>", # hide trace box
mode="markers",
marker=marker_options,
text=model_titles_reversed,
),
col=1,
row=1,
)
fig.add_trace(
go.Scatter(
x=alpha_norm_reversed,
y=alpha_theta_reversed,
hovertemplate="<b>%{text}</b><br>"
+ "|ɑ̅| = %{x:.3f}, "
+ "θ(ɑ̅) = %{y:.3f}"
+ "<extra></extra>", # hide trace box
mode="markers",
marker=marker_options,
text=model_titles_reversed,
),
col=2,
row=1,
)
fig.update_layout(
height=600,
showlegend=False,
title_text="Averaged ɑ̅ <b>systematics</b> distribution (polar)",
xaxis=dict(
title="ϕ(ɑ̅)",
tickmode="array",
tickvals=np.array([0.9, 0.95, 1, 1.05]) * np.pi,
ticktext=["0.9 π", "0.95 π", "π", "1.05 π"],
),
yaxis=dict(
title="θ(ɑ̅)",
tickmode="array",
tickvals=np.array([0.90, 0.91, 0.92, 0.93, 0.94, 0.95]) * np.pi,
ticktext=["0.90 π", "0.91 π", "0.92 π", "0.93 π", "0.94 π", "0.95 π"],
),
xaxis2=dict(title="|ɑ̅|"),
)
fig.show()
fig = go.Figure(
data=go.Scatter3d(
x=alpha_x_reversed,
y=alpha_y_reversed,
z=alpha_z_reversed,
hovertemplate="<b>%{text}</b><br>"
+ "ɑ̅<sub>x</sub> = %{x:.3f}, "
+ "ɑ̅<sub>y</sub> = %{y:.3f}, "
+ "ɑ̅<sub>z</sub> = %{z:.3f}"
+ "<extra></extra>", # hide trace box
mode="markers",
marker=marker_options,
text=model_titles_reversed,
),
)
fig.update_layout(
width=700,
height=700,
scene=dict(
aspectmode="cube",
xaxis_title="ɑ̅<sub>x</sub>",
yaxis_title="ɑ̅<sub>y</sub>",
zaxis_title="ɑ̅<sub>z</sub>",
),
showlegend=False,
title_text=R"Average ɑ̅ <b>systematics</b> distribution (cartesian)",
)
fig.show()
Show code cell source
def axis_formatter(decimals: int = 10):
# https://gist.github.com/taxus-d/aa69e43c3d8b804864ede4a8c056e9cd
def formatter(val, pos) -> str:
fraction = np.round(val / np.pi, decimals)
if fraction == 1:
return R"$\pi$"
return Rf"${fraction:g} \pi$"
return formatter
%config InlineBackend.figure_formats = ['svg']
plt.rcdefaults()
use_mpl_latex_fonts()
plt.rc("font", size=15)
fig, (ax1, ax2) = plt.subplots(figsize=(11, 5), ncols=2, sharey=True)
fig.suptitle(R"$\vec{\overline{\alpha}}$ statistics distribution (polar)", y=0.95)
fig.subplots_adjust(wspace=0.05)
norm, theta, phi = nominal_weighted_alpha_polar
ax1.set_xlabel(R"$\phi\left(\overline{\alpha}\right)$")
ax1.set_ylabel(R"$\theta\left(\overline{\alpha}\right)$")
ax2.set_xlabel(R"$\left|\overline{\alpha}\right|$")
ax1.xaxis.set_major_locator(MultipleLocator(base=0.05 * np.pi))
ax1.xaxis.set_major_formatter(FuncFormatter(axis_formatter(decimals=2)))
ax1.yaxis.set_major_locator(MultipleLocator(base=0.005 * np.pi))
ax1.yaxis.set_major_formatter(FuncFormatter(axis_formatter(decimals=3)))
ax1.scatter(
x=stat_weighted_alpha_polar[2], # phi
y=stat_weighted_alpha_polar[1], # theta
s=2,
)
ax1.scatter(phi, theta, c="red", marker="*")
ax1.annotate(
Rf"$\left({norm:.3f}, {theta/np.pi:+.3f}\pi, {phi/np.pi:+.3f}\pi\right)$",
xy=(phi, theta),
color="red",
fontsize=12,
)
ax2.scatter(
x=stat_weighted_alpha_polar[0], # norm
y=stat_weighted_alpha_polar[1], # theta
s=2,
label="Bootstraps",
)
ax2.scatter(norm, theta, c="red", marker="*", label="Nominal model")
ax2.annotate(
Rf"$\left({norm:.3f}, {theta/np.pi:+.3f}\pi, {phi/np.pi:+.3f}\pi\right)$",
xy=(norm, theta),
color="red",
fontsize=12,
)
ax2.legend(
bbox_to_anchor=(0.99, 0.18),
loc="upper right",
framealpha=1,
prop={"size": 12},
)
fig.savefig("_images/correlation-statistics.svg")
plt.show()
Show code cell source
%config InlineBackend.figure_formats = ['svg']
fig, (ax1, ax2) = plt.subplots(figsize=(11, 5), ncols=2, sharey=True)
fig.suptitle(R"$\vec{\overline{\alpha}}$ systematics distribution (polar)", y=0.95)
fig.subplots_adjust(wspace=0.05)
ax1.set_xlabel(R"$\phi\left(\overline{\alpha}\right)$")
ax1.set_ylabel(R"$\theta\left(\overline{\alpha}\right)$")
ax2.set_xlabel(R"$\left|\overline{\alpha}\right|$")
ax1.xaxis.set_major_locator(MultipleLocator(base=0.05 * np.pi))
ax1.xaxis.set_major_formatter(FuncFormatter(axis_formatter(decimals=2)))
ax1.yaxis.set_major_locator(MultipleLocator(base=0.01 * np.pi))
ax1.yaxis.set_major_formatter(FuncFormatter(axis_formatter(decimals=2)))
ax1.scatter(
x=syst_weighted_alpha_polar[2][1:],
y=syst_weighted_alpha_polar[1][1:],
marker=".",
)
ax1.scatter(phi, theta, c="red", marker="*")
ax1.annotate(
Rf"$\left({norm:.3f}, {theta/np.pi:+.3f}\pi, {phi/np.pi:+.3f}\pi\right)$",
xy=(phi, theta),
color="red",
fontsize=12,
)
ax2.scatter(
x=syst_weighted_alpha_polar[0][1:],
y=syst_weighted_alpha_polar[1][1:],
marker=".",
label="Alternative models",
)
ax2.scatter(norm, theta, c="red", marker="*", label="Nominal model")
ax2.annotate(
Rf"$\left({norm:.3f}, {theta/np.pi:+.3f}\pi, {phi/np.pi:+.3f}\pi\right)$",
xy=(norm, theta),
color="red",
fontsize=12,
)
ax2.legend(
bbox_to_anchor=(1.05, 1.1),
loc="upper right",
framealpha=1,
prop={"size": 12},
)
fig.savefig("_images/correlation-systematics.svg")
plt.show()
Show code cell source
def plot_correlation_matrix_polar(matrix, ax):
ax.set_box_aspect(1)
ax.matshow(matrix, cmap=cm.coolwarm, vmin=-1, vmax=+1)
for i, row in enumerate(matrix):
for j, value in enumerate(row):
ax.text(i, j, f"{value:.3f}", ha="center", va="center")
ticks = [i for i in range(3)]
tick_labels = [
R"$\left|\overline{\alpha}\right|$",
R"$\theta\left(\overline{\alpha}\right)$",
R"$\phi\left(\overline{\alpha}\right)$",
]
ax.set_xticks(ticks)
ax.set_xticklabels(tick_labels)
ax.set_yticks(ticks)
ax.set_yticklabels(tick_labels)
assert stat_weighted_alpha.shape == (3, n_bootstraps)
assert syst_weighted_alpha.shape == (3, n_models)
%config InlineBackend.figure_formats = ['svg']
fig, (ax1, ax2) = plt.subplots(figsize=(9, 5), ncols=2)
fig.suptitle(R"Correlation matrices for $\vec{\overline{\alpha}}$ (\textbf{polar})")
ax1.set_title(R"\textbf{statistics}")
ax2.set_title(R"\textbf{systematics}")
plot_correlation_matrix_polar(np.corrcoef(stat_weighted_alpha_polar), ax1)
plot_correlation_matrix_polar(np.corrcoef(syst_weighted_alpha_polar), ax2)
fig.savefig("_images/correlation-matrices.svg")
plt.show()
Show code cell source
def plot_correlation_matrix(matrix, ax):
ax.set_box_aspect(1)
ax.matshow(matrix, cmap=cm.coolwarm, vmin=-1, vmax=+1)
for i, row in enumerate(matrix):
for j, value in enumerate(row):
ax.text(i, j, f"{value:.3f}", ha="center", va="center")
ticks = [i for i in range(3)]
tick_labels = [Rf"$\overline{{\alpha}}_{i}$" for i in "xyz"]
ax.set_xticks(ticks)
ax.set_xticklabels(tick_labels)
ax.set_yticks(ticks)
ax.set_yticklabels(tick_labels)
assert stat_weighted_alpha.shape == (3, n_bootstraps)
assert syst_weighted_alpha.shape == (3, n_models)
%config InlineBackend.figure_formats = ['svg']
fig, (ax1, ax2) = plt.subplots(figsize=(9, 5), ncols=2)
fig.suptitle(
R"Correlation matrices for $\vec{\overline{\alpha}}$ (\textbf{cartesian})"
)
ax1.set_title(R"\textbf{statistical \& systematic}")
ax2.set_title(R"\textbf{model}")
plot_correlation_matrix(np.corrcoef(stat_weighted_alpha), ax1)
plot_correlation_matrix(np.corrcoef(syst_weighted_alpha), ax2)
fig.savefig("_images/correlation-matrices.svg")
plt.show()
Tip
A potential explanation for the \(xz\)-correlation may be found in Section XZ-correlations.
5.7. Exported distributions#
Export averaged polarimeter vectors
json_data = {
"file description": "Averaged polarimeter vector for each alternative model",
"model descriptions": list(models),
"reference subsystem": reference_subsystem,
"systematics": {
"cartesian": {
"x": syst_weighted_alpha[0].tolist(),
"y": syst_weighted_alpha[1].tolist(),
"z": syst_weighted_alpha[2].tolist(),
"norm": syst_weighted_alpha.tolist(),
},
"polar": {
"norm": syst_weighted_alpha_polar[0].tolist(),
"theta": syst_weighted_alpha_polar[1].tolist(),
"phi": syst_weighted_alpha_polar[2].tolist(),
},
},
"statistics": {
"cartesian": {
"x": stat_weighted_alpha[0].tolist(),
"y": stat_weighted_alpha[1].tolist(),
"z": stat_weighted_alpha[2].tolist(),
"norm": stat_weighted_alpha.tolist(),
},
"polar": {
"norm": stat_weighted_alpha_polar[0].tolist(),
"theta": stat_weighted_alpha_polar[1].tolist(),
"phi": stat_weighted_alpha_polar[2].tolist(),
},
},
}
json_averaged = "_static/export/averaged-polarimeter-vectors.json"
with open(json_averaged, "w") as f:
json.dump(json_data, f, indent=2)
del json_data
Define Dalitz grid
grid_resolution = 100
grid_sample = generate_meshgrid_sample(model.decay, grid_resolution)
grid_sample = transformer(grid_sample)
X_export = grid_sample["sigma1"]
Y_export = grid_sample["sigma2"]
Export fields as JSON
def format_subsystem(reference_subsystem) -> str:
subsystem_names = {
1: R"K^{**} \to \pi^+ K^-",
2: R"\Lambda^{**} \to p K^-",
3: R"\Delta^{**} \to p \pi^+",
}
name = subsystem_names[reference_subsystem]
return f"{reference_subsystem}: {name}"
STAT_FILENAMES = []
for i in tqdm(range(n_bootstraps), disable=NO_TQDM):
new_parameters = {k: v[i] for k, v in bootstrap_parameters.items()}
for key, func in nominal_functions.items():
func.update_parameters(nominal_parameters)
func.update_parameters(new_parameters)
filename = f"_static/export/polarimetry-field-bootstrap-{i}.json"
export_polarimetry_field(
sigma1=X_export[0],
sigma2=Y_export[:, 0],
intensity=nominal_functions["intensity"](grid_sample).real,
alpha_x=nominal_functions["alpha_x"](grid_sample).real,
alpha_y=nominal_functions["alpha_y"](grid_sample).real,
alpha_z=nominal_functions["alpha_z"](grid_sample).real,
filename=filename,
metadata={
"model description": nominal_model_title,
"parameters": {k: f"{v}" for k, v in new_parameters.items()},
"reference subsystem": format_subsystem(reference_subsystem),
},
)
STAT_FILENAMES.append(filename)
items = list(enumerate(jax_functions.items()))
SYST_FILENAMES = []
for i, (title, funcs) in tqdm(items, disable=NO_TQDM):
for func in funcs.values():
func.update_parameters(original_parameters[title])
filename = f"_static/export/polarimetry-field-model-{i}.json"
export_polarimetry_field(
sigma1=X_export[0],
sigma2=Y_export[:, 0],
intensity=funcs["intensity"](grid_sample).real,
alpha_x=funcs["alpha_x"](grid_sample).real,
alpha_y=funcs["alpha_y"](grid_sample).real,
alpha_z=funcs["alpha_z"](grid_sample).real,
filename=filename,
metadata={
"model description": title,
"parameters": {k: f"{v}" for k, v in func.parameters.items()},
"reference subsystem": format_subsystem(reference_subsystem),
},
)
SYST_FILENAMES.append(filename)
Merge into one TAR/JSON file
models_key = "models"
bootstraps_key = "bootstraps"
s1_key = "m^2_Kpi"
s2_key = "m^2_pK"
combined_json = {
models_key: [],
bootstraps_key: [],
}
for filename in SYST_FILENAMES:
with open(filename) as f:
data = json.load(f)
s1_values = data[s1_key]
s2_values = data[s2_key]
del data[s1_key]
del data[s2_key]
combined_json[models_key].append(data)
for filename in STAT_FILENAMES:
with open(filename) as f:
data = json.load(f)
del data[s1_key]
del data[s2_key]
combined_json[bootstraps_key].append(data)
combined_json[s1_key] = s1_values
combined_json[s2_key] = s2_values
json_file = "_static/export/polarimetry-field.json"
with open(json_file, "w") as f:
json.dump(combined_json, f)
tar_file = "_static/export/polarimetry-field.tar.gz"
with tarfile.open(tar_file, "w:gz") as tar:
tar.add("_static/export/README.md", arcname="README.md")
for path in STAT_FILENAMES + SYST_FILENAMES:
tar.add(path, arcname=os.path.basename(path))
Exported 100x100 JSON grids for each bootstrap (statistics & systematics)
Exported 100x100 JSON grids for each model
[download] Default amplitude model
[download] Alternative amplitude model with K(892) with free mass and width
[download] Alternative amplitude model with L(1670) with free mass and width
[download] Alternative amplitude model with L(1690) with free mass and width
[download] Alternative amplitude model with D(1232) with free mass and width
[download] Alternative amplitude model with L(1600), D(1600), D(1700) with free mass and width
[download] Alternative amplitude model with free L(1405) Flatt’e widths, indicated as G1 (pK channel) and G2 (Sigmapi)
[download] Alternative amplitude model with L(1800) contribution added with free mass and width
[download] Alternative amplitude model with L(1810) contribution added with free mass and width
[download] Alternative amplitude model with D(1620) contribution added with free mass and width
[download] Alternative amplitude model in which a Relativistic Breit-Wigner is used for the K(700) contribution
[download] Alternative amplitude model with K(700) with free mass and width
[download] Alternative amplitude model with K(1410) contribution added with mass and width from PDG2020
[download] Alternative amplitude model in which a Relativistic Breit-Wigner is used for the K(1430) contribution
[download] Alternative amplitude model with K(1430) with free width
[download] Alternative amplitude model with an additional overall exponential form factor exp(-alpha q^2) multiplying Bugg lineshapes. The exponential parameter is indicated as ``alpha’’
[download] Alternative amplitude model with free radial parameter d for the Lc resonance, indicated as dLc
[download] Alternative amplitude model obtained using LS couplings
averaged-polarimeter-vectors.json (34.0 kB)
Tip
See Import and interpolate for how to use these grids in an an analysis and see Determination of polarization for how to use these fields to determine the polarization from a measured distribution.
